Overview
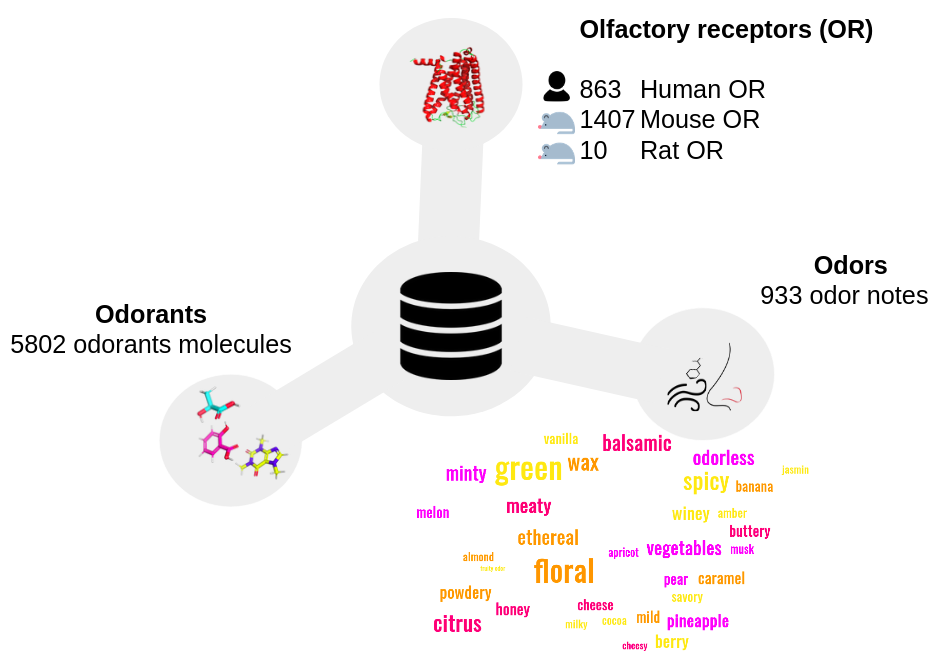
Pred-O3 is a web server that compiles information about odorant molecules, their odors and their olfactory receptors (OR). The server integrates several tools to analyze the interactions between odorants, olfactory receptors and their physico-chemical properties. We freely give access to a compiled database of 5802 chemicals with known odors. The database contains 385 olfactory receptors (from human, mouse and rat species) for which at least one chemical-or interaction is known. There are 36016 known odorant-odor links and 2732 known odorant-OR interactions. The website proposes to search for olfactory receptors and/or odor annotated for a chemical (using its name or chemical structure). Also, through several tools, Pred-O3 gives the opportunity to predict olfactory receptors and odors associated to any small molecules using a graphical neural network methodology (GNN). Finally, the webserver predicts the ligand-olfactory receptor binding mode for a set of 1572 ORs.
Tools
Structural similarity
Similar compounds to the proposed database can be searched using by 2 methods : 1- Similarity search using MACC fingerprints and a Tanimoto metric 2- Substructure search using structural pattern of an odorant molecule can be searched.
Prediction
Predict the putative odors of a molecule or its potential interacting olfactory receptors, using two developed models based on a Graphical Neural Network (GNN) developed by Achebouche et al.
Docking
Select a chemical and an olfactory receptor and launch the SeamDock webserver [Murail et al] to predict ligand-OR binding mode.
References
Ollitrault G, Achebouche R, Dreux A, Murail S, Audouze K, Tromelin A, Taboureau O.
Pred-O3, a web server to predict molecules, olfactory receptors and odor relationships. Nucleic Acids Res. (2024).
https://doi.org/10.1093/nar/gkae305
Achebouche R, Tromelin A, Audouze K & Taboureau O.
Application of artificial intelligence to decode the relationships between smell, olfactory receptors and small molecules. Sci Rep 12, 18817 (2022).
https://doi.org/10.1038/s41598-022-23176-y
Murail S, de Vries S, Rey J, Moroy G & Tufféry P.
SeamDock: An Interactive and Collaborative Online Docking Resource to Assist Small Compound Molecular Docking. Frontiers in Molecular Biosciences (2021).
https://doi.org/10.3389/fmolb.2021.716466
Tufféry P & Murail S.
Docking_py, a python library for ligand protein docking. Zenodo (2020).
http://doi.org/10.5281/zenodo.4506970.