Structure search
Structural similarity search can be performed by either searching for overall similar structure though a tanimoto score or by searching for sub-structures.
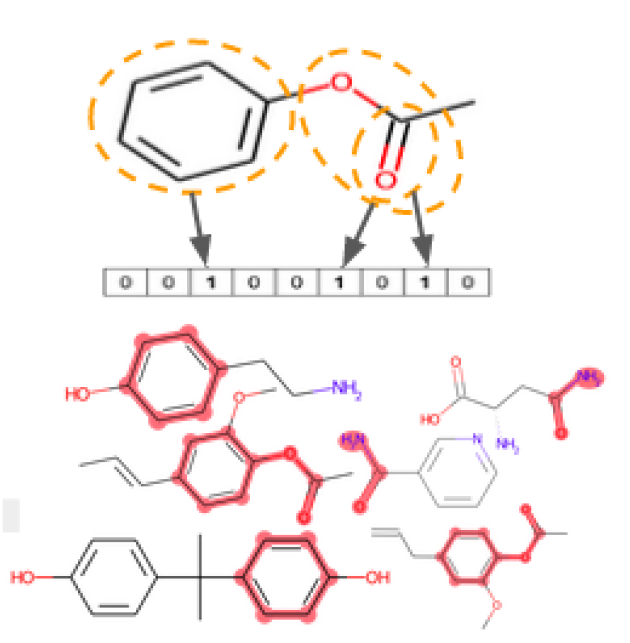
The similarity search based on a tanimoto score for a query molecule returns a score between 0 and 1 (similar) for all the molecules in the database. The molecule encodes MACCS fingerprint which is a bit vector of 120 bit counting the presence (1) or absence (0) of a specific functional group in the structure of the molecule. The tanimoto score varies between 0 and 1 the higher the score the higher the similarity between the molecules. Molecules are then sorted from most similar to the lowest.
The substructure search where a substructure pattern of an odorant molecule can be searched via a smart pattern or any molecule identifier available in the searching method. It returns all the molecules of the database with the query substructure in its structure.
Draw structure/substructure:
Input structure/substructure:
